Today, we’re announcing research on one of the industry’s first full-body 3D Magnetic Resonance Imaging (MRI) foundation models, designed to enable developers to build applications for tasks such as image retrieval, classification, image segmentation, and report generation. With this new multipurpose, multimodal model, developers could build applications that allow clinicians to interact with full 3D organ models in a single computational pass.
This model showed up to 30% accuracy in matching MRI scans with textual descriptions in image retrieval tasks — a significant improvement over the 3% capability demonstrated by similar models like Biomed CLIP¹. Additionally, in internal testing, the model achieved full performance level in disease detection (classification of images to reports) within just ten training cycles, compared to fifty or more epochs required by previous models. This can enable healthcare application developers to conduct more experiments with the same resources or reduce training time and associated costs by up to 5X.
The foundation model introduced today is among the industry’s first to bring 3D capabilities to MR image processing and analysis. Prior 2D MRI AI models can limit the ability to analyze intricate anatomical structures, especially in complex cases such as brain tumors, cardiovascular disease, and musculoskeletal disorders. This 3D MRI foundation model is designed to overcome these limitations by transforming 2D MRI slices into fully detailed 3D body representations.
Our vision is for the model to align seamlessly with established clinical reporting methodologies like BI-RADS (Breast Imaging Reporting and Data System), to help enhance the objectivity, consistency, and efficiency of breast cancer diagnosis and risk stratification. The model builds on an inherent advantage: 3D MRIs can capture comprehensive anatomical and pathological details, which can allow for better characterization of breast lesions (such as size, shape, margins, and internal characteristics). The model can learn to recognize specific patterns such as irregular borders and rapid contrast uptake that correlate with higher BI-RADS categories (e.g., BI-RADS 4 or 5).
Compared to other publicly available foundation models, this model demonstrated enhanced performance on tasks² like image retrieval, classification, and grounding, due to its MRI-specific dataset, advanced 3D functionality, and specialized architecture and pre-training methods.
We developed this technology by training the first generation of the model on a dataset of more than 173,000 MRI images from more than 19,000 studies, using only MRI images. This exclusive focus on MRI improved the model’s accuracy in tasks such as prostate cancer and Alzheimer’s disease classification, even when data for fine-tuning is limited. Additionally, this model was trained on data originating from various MRI techniques, thus giving the model the ability to identify distinct features of soft tissues like the brain, muscles, and organs across MRI techniques, such as T1 for soft-tissue anatomy and fat, T2 for fluid, and diffusion-weighted imaging for detecting restricted water movement, each highlighting unique aspects of the body's internal structures.
This MRI-centric training sets it apart from foundation models designed for X-rays and CT scans, which rely on X-rays to image bones and dense structures, whereas MRI uses magnets to capture detailed soft tissue features essential for its imaging. The model’s architecture and training techniques are designed to boost performance across a range of tasks and use cases.
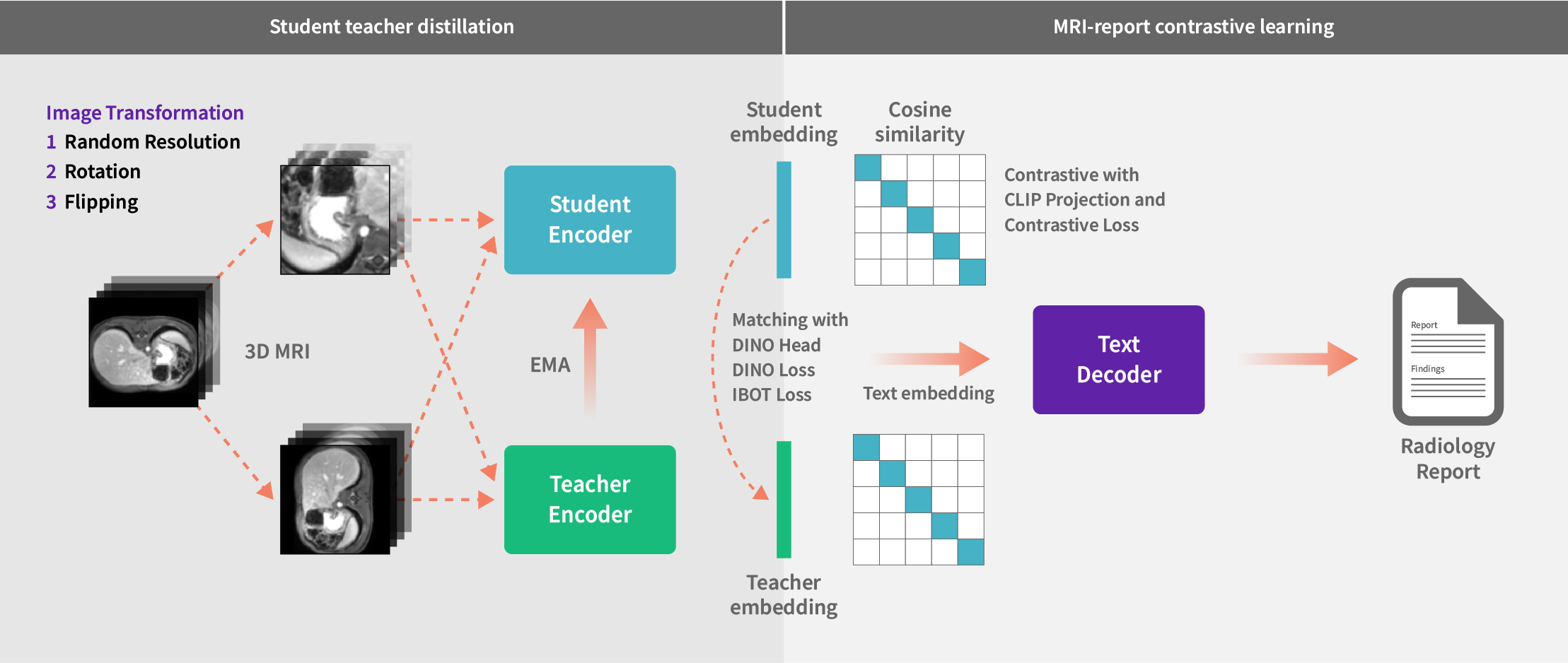
Using student-teacher distillation and contrastive learning to link 3D MRI images with radiology reports
By leveraging this extensive and diverse dataset, we pretrained our foundation model using a combination of state-of-the-art student-teacher self-supervised learning (a technique where a smaller "student" model learns from a larger "teacher" model without requiring labeled data) and vision-language contrastive learning (a method that aligns visual and textual data by teaching the model to associate images with their corresponding text and distinguish them from unrelated pairs). These approaches are known to enhance the model’s accuracy, reduce dependence on labeled data, integrate multimodal insights, and could potentially streamline workflows to offer healthcare institutions a scalable, cost-efficient solution for improved patient care.
We utilized a range of Vision Transformer models tailored to varying computational capacities. The base model strikes a balance between performance and resource efficiency, ideal for environments with moderate computational power. In addition, advanced models featuring deeper layers and extensive attention mechanisms can be leveraged to enable superior performance on tasks like image retrieval from text prompts and disease detection.
To maximize the use of available data during pretraining, we refined our strategy to handle missing reports by masking absent text during backpropagation, enabling the model to learn effectively from existing MRI information. Additionally, we implemented an innovative data augmentation strategy, allowing the model to process MRI scans of varying resolutions and sizes, ensuring robust performance across diverse data characteristics. This adaptability is particularly valuable in resource-constrained settings, highlighting progress towards a model ready for real-world deployment where consistent and generalizable performance is essential.
Our 3D MRI foundation model was tested across multiple benchmarking tasks, demonstrating flexibility and potential to enhance MRI analysis. Early results indicated improvements across key performance metrics for tasks related to classification, image retrieval, segmentation, and report generation – detailed results are provided in each of the sections below.
Classification
The model demonstrated exceptional performance in differentiating normal from disease cases on a public dataset containing Alzheimer’s, prostate cancer, or cardiac conditions, surpassing existing foundation models in accuracy. On classification benchmarks across body regions—such as detecting Alzheimer’s, cardiac diseases, and prostate cancer—our model achieved an average 8% improvement over the state-of-the-art BioMed CLIP model. Its versatility across various body regions could offer an opportunity to build solutions for various care pathways. Notably, the model reached peak accuracy within just ten training epochs, requiring no specialized data preparation. This efficiency makes it likely ideal for training on new tasks, reducing costs and boosting performance in downstream applications.
Image Retrieval
In image retrieval tests—assessing the ability to retrieve an image from a text prompt—the model excelled at matching MRI scans with corresponding text descriptions. Using an in-domain dataset comprising 25,000 MRI images from 2,500 studies, which included diverse body regions, MR sequences, and disease conditions, the model achieved nearly 30% success in retrieving the original scan within the top ten results based on short report phrases (e.g., “MRI with organ name/tumor type”). This performance vastly outpaced BioMed CLIP, which achieved only 3% under similar conditions. Once developed, this capability can help clinicians retrieve comparable cases more efficiently.
Segmentation
For organ segmentation in radiation therapy planning—where the goal is to produce precise outlines of organ contours—the model demonstrated significant improvements in accuracy, achieving markedly higher Dice scores compared to prior models. By leveraging our MR model as an encoder for feature extraction and pairing it with a customized decoder for segmentation, the model delivered 10% to 40% greater accuracy in segmenting head, neck, and abdomen organs compared to the encoder from the publicly available SAM-Med 3D model.
Grounding
For phrase grounding—the ability to identify and localize specific entities within medical images based on a phrase query—the model outperformed the encoder from the DETR3D model, locating head and neck organs with 25% greater accuracy. Additionally, on a curated dataset featuring cases with missing or partially absent organs and various tumors, our model delivered 9% better performance than DETR3D. This capability could enhance the interpretability of AI-generated insights, by linking reports to precise visual findings and areas of interest, thus helping foster trust in the model's diagnostic support.
Report Generation
We are evaluating the model's performance in automating MRI report creation, a capability with the potential to significantly reduce radiologists' workloads by generating comprehensive reports directly from MRI images.
At GE HealthCare, we are committed to the deployment of AI in a responsible way and are exploring a number of techniques to manage the risks associated with generative AI. To give just one example of how we are addressing these risks, we are building capability for visual grounding to improve image understanding by allowing for more precise identification of findings.
For instance, take the example of a radiologist generating a report describing conditions observed in an MRI, such as brain lesions or soft tissue abnormalities. The report would then be processed by a phrase grounding system, which highlights specific areas on the MRI that correspond to the descriptions, to assist the radiologist by visually marking relevant regions and ensuring consistency in interpretation.
Our 3D MRI foundation model offers a groundbreaking approach to medical imaging analysis and research, designed to capture the intricate complexity of 3D MRI data. As demonstrated above, the model outperforms other publicly available research models in tasks such as classification, segmentation, and image retrieval. Its flexibility and reliability can make it an invaluable resource for researchers, developers, and clinicians to help pave the way for improved patient care. We encourage the community to connect with us and explore how this model can advance your research and clinical applications.
¹ Biomed research showing data exposure available at https://arxiv.org/pdf/2303.00915
² Data on file
Concept only. This work is in concept phase and may never become a product. Not for Sale. Any reported results are preliminary and subject to change. Not cleared or approved by the U.S. FDA or any other global regulator for commercial availability.